Publications
Some of our recent publications are included below. You can find a more comprehensive, up to date list at Google Scholar.
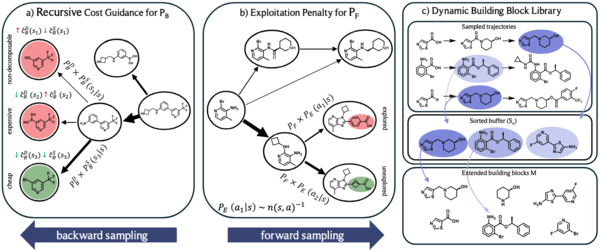
Scalable and cost-efficient de novo template-based molecular generation
Gaiński, P., Boussif, O., Shevchuk, D., Rekesh, A., Parviz, A., Tyers, M., Batey, R.A., Koziarski, M.
NeurIPS. 2025.
Link to paper
Gaiński, P., Boussif, O., Shevchuk, D., Rekesh, A., Parviz, A., Tyers, M., Batey, R.A., Koziarski, M.
NeurIPS. 2025.
Link to paper
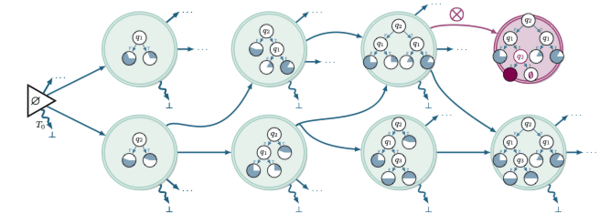
Learning decision trees as amortized structure inference
Mahfoud, M., Boukachab, G., Koziarski, M., Hernandez-Garcia, A., Bauer, S., Bengio, Y., Malkin, N.
arXiv. 2025.
Link to paper
Mahfoud, M., Boukachab, G., Koziarski, M., Hernandez-Garcia, A., Bauer, S., Bengio, Y., Malkin, N.
arXiv. 2025.
Link to paper
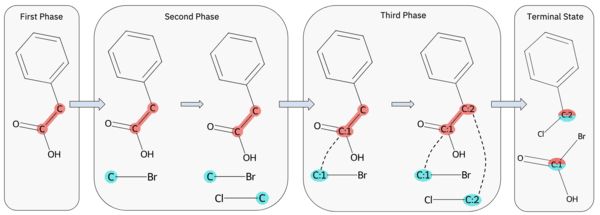
Diverse and feasible retrosynthesis using GFlowNets
Gaiński, P., Koziarski, M., Maziarz, K., Segler, M., Tabor, J. and Śmieja, M.
Information Sciences. 2025.
Link to paper
Gaiński, P., Koziarski, M., Maziarz, K., Segler, M., Tabor, J. and Śmieja, M.
Information Sciences. 2025.
Link to paper
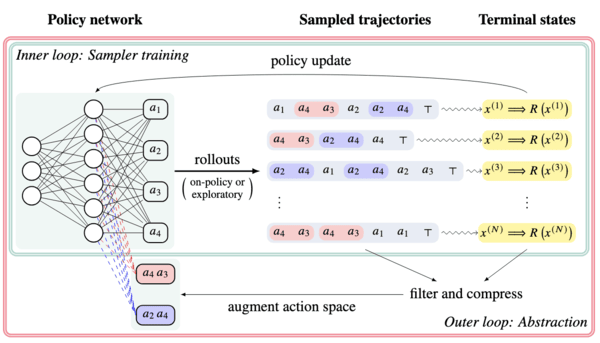
Action abstractions for amortized sampling
Boussif, O., Ezzine, L.N., Viviano, J.D., Koziarski, M., Jain, M., Malkin, N., Bengio, E., Assouel, R., Bengio, Y.
ICLR. 2025.
Link to paper
Boussif, O., Ezzine, L.N., Viviano, J.D., Koziarski, M., Jain, M., Malkin, N., Bengio, E., Assouel, R., Bengio, Y.
ICLR. 2025.
Link to paper
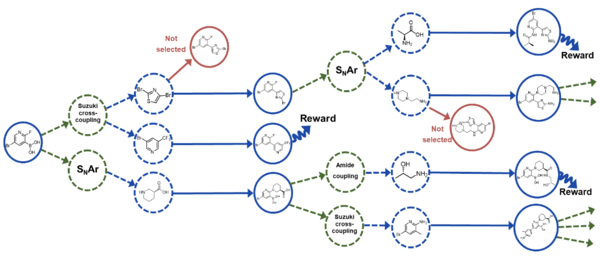
RGFN: synthesizable molecular generation using GFlowNets
Koziarski, M., Rekesh, A., Shevchuk, D., van der Sloot, A., Gaiński, P., Bengio, Y., Liu, C.H., Tyers, M. and Batey, R.A.
NeurIPS. 2024.
Link to paper
Koziarski, M., Rekesh, A., Shevchuk, D., van der Sloot, A., Gaiński, P., Bengio, Y., Liu, C.H., Tyers, M. and Batey, R.A.
NeurIPS. 2024.
Link to paper
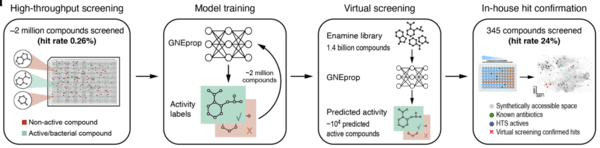
A high-throughput phenotypic screen combined with an ultra-large-scale deep learning-based virtual screening reveals novel scaffolds of antibacterial compounds
Scalia, G., Rutherford, S. T., Lu, Z., Buchholz, K. R., Skelton, N., Chuang, K., ... & Biancalani, T.
bioRxiv. 2024.
Link to paper
Scalia, G., Rutherford, S. T., Lu, Z., Buchholz, K. R., Skelton, N., Chuang, K., ... & Biancalani, T.
bioRxiv. 2024.
Link to paper
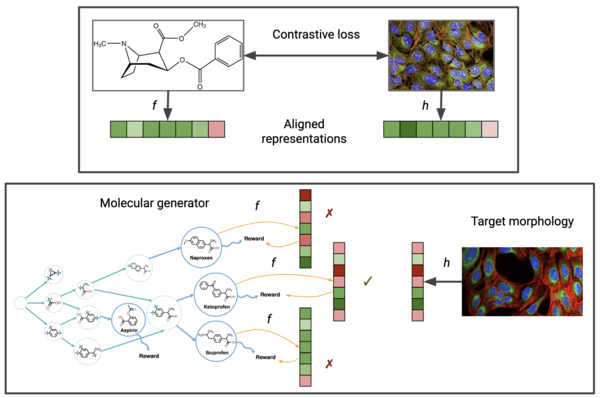
Cell morphology-guided small molecule generation with GFlowNets
Lu, S.Z., Lu, Z., Hajiramezanali, E., Biancalani, T., Bengio, Y., Scalia, G. and Koziarski, M.
ICML AI for Science Workshop. 2024.
Link to paper
Lu, S.Z., Lu, Z., Hajiramezanali, E., Biancalani, T., Bengio, Y., Scalia, G. and Koziarski, M.
ICML AI for Science Workshop. 2024.
Link to paper
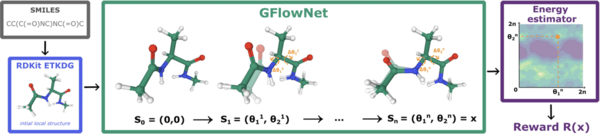
Towards equilibrium molecular conformation generation with GFlowNets
Volokhova, A., Koziarski, M., Hernández-García, A., Liu, C.H., Miret, S., Lemos, P., Thiede, L., Yan, Z., Aspuru-Guzik, A. and Bengio, Y.
Digital Discovery. 2024.
Link to paper
Volokhova, A., Koziarski, M., Hernández-García, A., Liu, C.H., Miret, S., Lemos, P., Thiede, L., Yan, Z., Aspuru-Guzik, A. and Bengio, Y.
Digital Discovery. 2024.
Link to paper
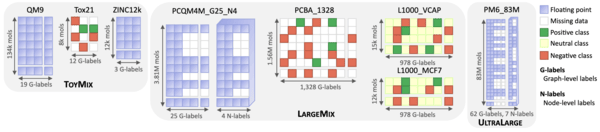
Towards foundational models for molecular learning on large-scale multi-task datasets
Beaini, D., Huang, S., Cunha, J. A., Li, Z., Moisescu-Pareja, G., Dymov, O., ... & Masters, D.
ICLR. 2024.
Link to paper
Beaini, D., Huang, S., Cunha, J. A., Li, Z., Moisescu-Pareja, G., Dymov, O., ... & Masters, D.
ICLR. 2024.
Link to paper
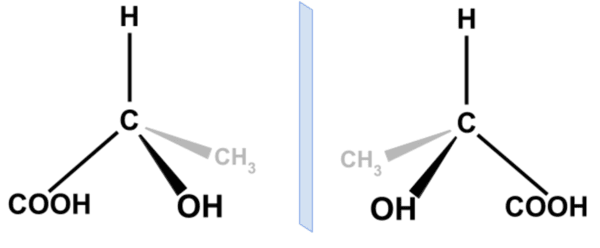
ChiENN: embracing molecular chirality with graph neural networks
Gaiński, P., Koziarski, M., Tabor, J. and Śmieja, M.
ECML. 2023.
Link to paper
Gaiński, P., Koziarski, M., Tabor, J. and Śmieja, M.
ECML. 2023.
Link to paper